These graphs have been generated by the PIGOKsum CGI or the PIGOKlookup CGI.
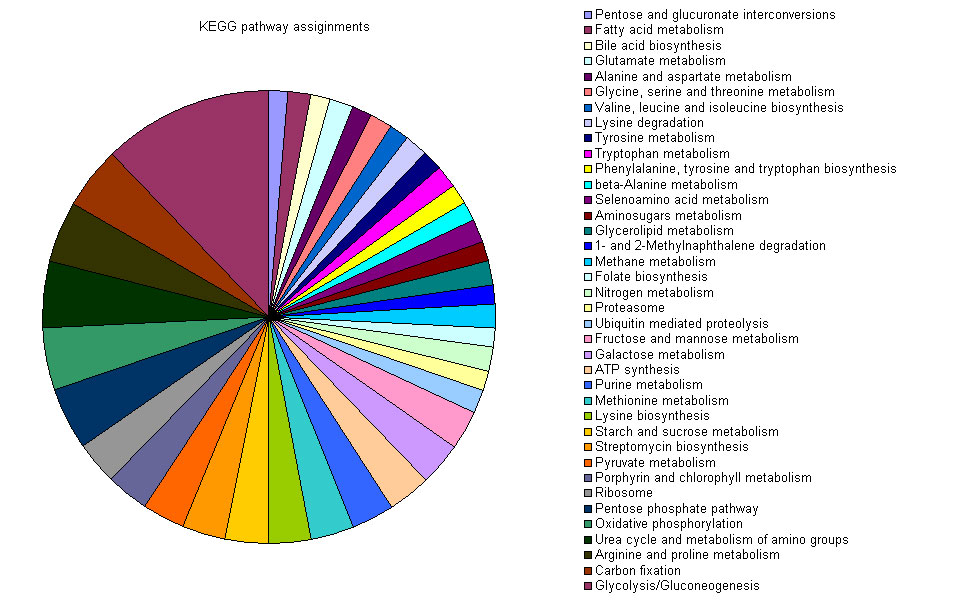
The Codon Adaptation Index(CAI) has been show to have a good correlation with
gene expression levels[ref]. Here I compare the CAI profile between the whole genome and the oxidative stress induced up and down regulated proteins identified from a S. pombe 2D gel[ref].
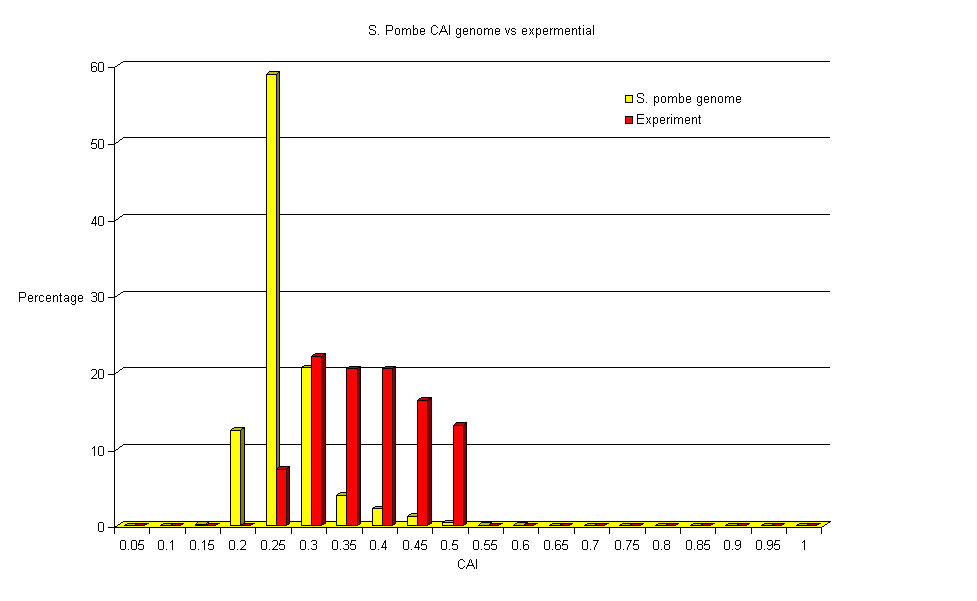
I have used the same data set to generate this Gene Ontology pie chart[ref].
The inner ring is the GO Slim biological process values for the whole S. pombe geneome
compared to the outer ring of the identified up and down regulated proteins.
Our identified proteins have a higher proportion of proteins that are involved in the response to stress and the generation of precursor metabolites and energy as might be expected for cells under stress.
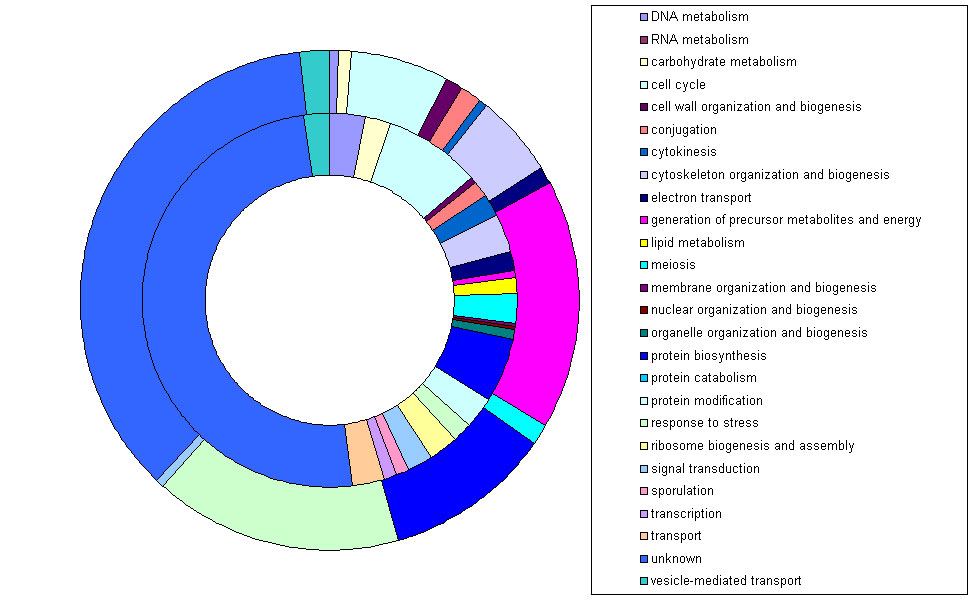
Finally I used the optional Kyoto Encyclopedia of Genes and Genomes(KEGG) direct link feature to map the identified proteins on to KEGG pathways. Only 54 proteins from the
data set could be mapped to KEGG pathways but they were involved in 39 different pathways with 8 proteins been identified from the Glycolysis/Gluconeogenesis pathway.